Methods¶
Permutation Importance¶
Theory¶
Permutation importance, the namesake for this package, determines the predictors which are important by comparing the performance of a model on a dataset where some of the predictors are individually permuted to performance on the original dataset without any permutation. The permutation of the individual predictor in this manner effectively breaks the relationship between the input predictor and the target variable. The predictor which, when permuted, results in the worst performance is typically taken as the most important variable. Permutation importance has the distinct advantage of not needing to retrain the model each time.
This method was originally designed for random forests by Breiman (2001), but can be used by any model. The original version of the algorithm was 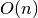 , but this was later revised by Lakshmanan (2015) to be more robust to correlated predictors and is
, but this was later revised by Lakshmanan (2015) to be more robust to correlated predictors and is 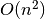 . The difference between these two methods is detailed in Fig. 1: Singlepass permutation importance and Fig. 2: Multipass permutation importance. While Breiman’s method only permutes each predictor once independently, Lakshmanan’s method iteratively adds one predictor to the set of predictors which are permuted at each iteration. Because Lakshmanan’s method can be viewed as successively applying Breiman’s method to determine the next-most important predictor, we typically refer to Breiman’s method as “singlepass” and Lakshmanan’s method as “multipass”.
. The difference between these two methods is detailed in Fig. 1: Singlepass permutation importance and Fig. 2: Multipass permutation importance. While Breiman’s method only permutes each predictor once independently, Lakshmanan’s method iteratively adds one predictor to the set of predictors which are permuted at each iteration. Because Lakshmanan’s method can be viewed as successively applying Breiman’s method to determine the next-most important predictor, we typically refer to Breiman’s method as “singlepass” and Lakshmanan’s method as “multipass”.
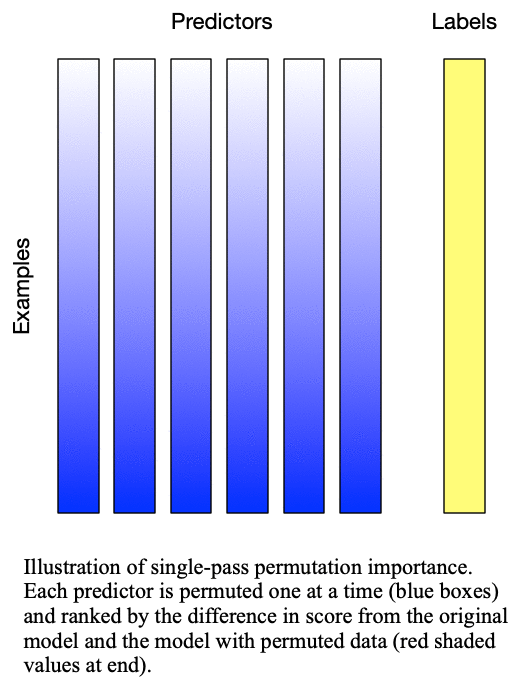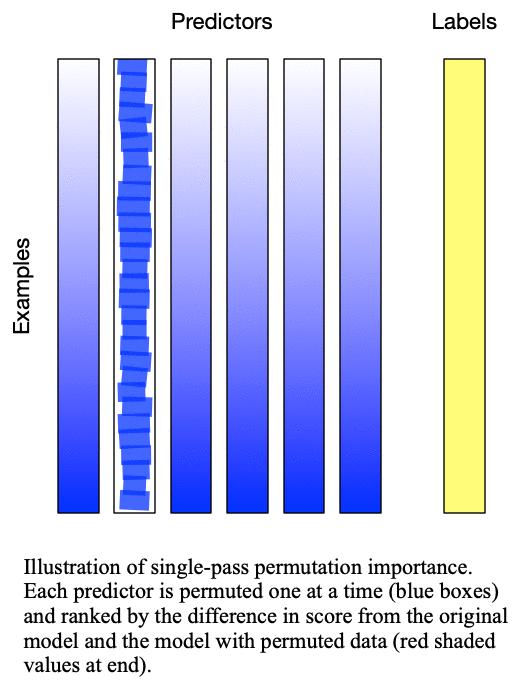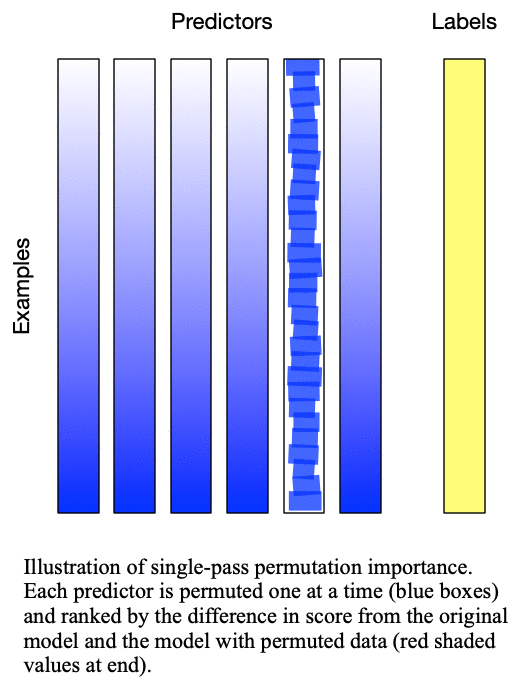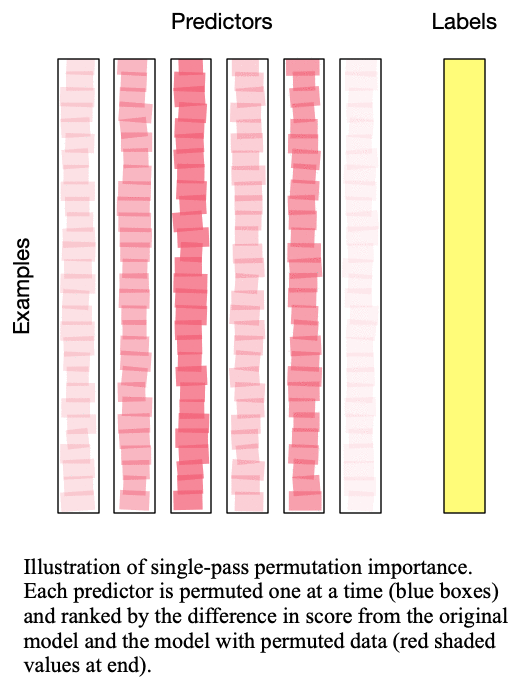
Fig. 1: Singlepass permutation importance evaluates each predictor independently by permuting only the values of that predictor
Fig. 2: Multipass permutation importance performs singlepass permutation importance as many times as there as predictors to iteratively determine the next-most important predictor
Usage¶
As with all methods, we provide the permutation importance method at two different levels of abstraction. For more information on the levels of abstraction and when to use each, please see Levels of Abstraction.
Singlepass permutation importance is computed as a byproduct of the generalized method. To compute singlepass permutation importance only, set nimportant_vars=1, which will only perform the multipass method for precisely one pass.
Typically, when using a performance metric or skill score with permutation
importance, the scoring_strategy should be to minimize the performance. On the
other hand, when using an error or loss function, the scoring_strategy should
be to maximize the error or loss function.
Model-Based¶
-
PermutationImportance.permutation_importance.sklearn_permutation_importance(model, scoring_data, evaluation_fn, scoring_strategy, variable_names=None, nimportant_vars=None, njobs=1, nbootstrap=1, subsample=1, **kwargs)[source] Performs permutation importance for a particular model,
scoring_data,evaluation_fn, and strategy for determining optimal variablesParameters: - model – a trained sklearn model
- scoring_data – a 2-tuple
(inputs, outputs)for scoring in thescoring_fn - evaluation_fn – a function which takes the deterministic or
probabilistic model predictions and scores them against the true
values. Must be of the form
(truths, predictions) -> some_valueProbably one of the metrics inPermutationImportance.metricsor sklearn.metrics - scoring_strategy – a function to be used for determining optimal
variables. Should be of the form
([some_value]) -> index - variable_names – an optional list for variable names. If not given, will use names of columns of data (if pandas dataframe) or column indices
- nimportant_vars – number of variables to compute multipass importance for. Defaults to all variables
- njobs – an integer for the number of threads to use. If negative, will
use
num_cpus + njobs. Defaults to 1 - nbootstrap – number of times to perform scoring on each variable. Results over different bootstrap iterations are averaged. Defaults to 1
- subsample – number of elements to sample (with replacement) per bootstrap round. If between 0 and 1, treated as a fraction of the number of total number of events (e.g. 0.5 means half the number of events). If not specified, subsampling will not be used and the entire data will be used (without replacement)
- kwargs – all other kwargs will be passed on to the
evaluation_fn
Returns: PermutationImportance.result.ImportanceResultobject which contains the results for each run
Simple Example¶
from sklearn.datasets import load_iris
from sklearn.metrics import accuracy_score
from sklearn.neural_network import MLPClassifier
from PermutationImportance import sklearn_permutation_importance
# Separate out the last 20% for scoring data
iris = load_iris(return_X_y=False)
inputs = iris.get('data')
outputs = iris.get('target')
predictor_names = iris.get('feature_names')
training_inputs = inputs[:int(0.8 * len(inputs))]
training_outputs = outputs[:int(0.8 * len(outputs))]
scoring_inputs = inputs[int(0.8 * len(inputs)):]
scoring_outputs = outputs[int(0.8 * len(outputs)):]
# Train a quick neural net on the data
model = MLPClassifier(solver='lbfgs')
model.fit(training_inputs, training_outputs)
# Package the data into the right shape
scoring_data = (scoring_inputs, scoring_outputs)
# Use the sklearn_permutation_importance to compute importances
result = sklearn_permutation_importance(
model, scoring_data, accuracy_score, 'min', variable_names=predictor_names)
# Get the Breiman-like singlepass results
print("Singlepass")
singlepass = result.retrieve_singlepass()
for predictor in singlepass.keys():
rank, score = singlepass[predictor]
print("Predictor: %s, Rank: %i, Score: %f" % (predictor, rank, score))
# Get the Lakshmanan-like multipass results
print("Multipass")
multipass = result.retrieve_multipass()
for predictor in multipass.keys():
rank, score = multipass[predictor]
print("Predictor: %s, Rank: %i, Score: %f" % (predictor, rank, score))
# Iterate over the (context, result) pairs
for i, (cntxt, res) in enumerate(result):
print("Context %i: %r" % (i, cntxt))
print("Result %i: %r" % (i, res))
Complex Example¶
from sklearn.datasets import load_breast_cancer
from sklearn.ensemble import RandomForestClassifier
from PermutationImportance import sklearn_permutation_importance
from PermutationImportance.metrics import peirce_skill_score
# Separate out the last 20% for scoring data
breast_cancer = load_breast_cancer(return_X_y=False)
inputs = breast_cancer.get('data')
outputs = breast_cancer.get('target')
predictor_names = breast_cancer.get('feature_names')
training_inputs = inputs[:int(0.8 * len(inputs))]
training_outputs = outputs[:int(0.8 * len(outputs))]
scoring_inputs = inputs[int(0.8 * len(inputs)):]
scoring_outputs = outputs[int(0.8 * len(outputs)):]
# Train a quick forest on the data
model = RandomForestClassifier(n_estimators=100, max_depth=4)
model.fit(training_inputs, training_outputs)
# Package the data into the right shape
scoring_data = (scoring_inputs, scoring_outputs)
# ----------- Version to use when only wanting singlepass results --------------
# Use the sklearn_permutation_importance to compute importances
result = sklearn_permutation_importance(
# argmin_of_mean handles bootstrapped metrics
model, scoring_data, peirce_skill_score, 'argmin_of_mean',
variable_names=predictor_names,
# sample (with replacement) 1*(number of samples) 5 times to compute metric distribution
# nbootstrap should typically be 1000, but this is kept small here for printing purposes
nbootstrap=5, subsample=1,
# only perform for the very top predictor (effectively means only compute singlepass results)
nimportant_vars=1)
# Get the Breiman-like singlepass results
print("Singlepass")
singlepass = result.retrieve_singlepass()
for predictor in singlepass.keys():
rank, score = singlepass[predictor]
print("Predictor: %s, Rank: %i, Score: %r" % (predictor, rank, score))
# Get the Lakshmanan-like multipass results
print("Multipass. This should only have 1 item and be not very useful")
multipass = result.retrieve_multipass()
for predictor in multipass.keys():
rank, score = multipass[predictor]
print("Predictor: %s, Rank: %i, Score: %r" % (predictor, rank, score))
# Iterate over the (context, result) pairs
for i, (cntxt, res) in enumerate(result):
print("Context %i: %r" % (i, cntxt))
print("Result %i: %r" % (i, res))
# ------------------------------------------------------------------------------
# ----------- Version to use when wanting multipass results --------------------
# Use the sklearn_permutation_importance to compute importances
result = sklearn_permutation_importance(
# argmin_of_mean handles bootstrapped metrics
model, scoring_data, peirce_skill_score, 'argmin_of_mean',
variable_names=predictor_names,
# sample (with replacement) 1*(number of samples) 5 times to compute metric distribution
# nbootstrap should typically be 1000, but this is kept small here for printing purposes
nbootstrap=5, subsample=1,
nimportant_vars=8) # only perform for the top 8 predictors
# Get the Breiman-like singlepass results
print("Singlepass")
singlepass = result.retrieve_singlepass()
for predictor in singlepass.keys():
rank, score = singlepass[predictor]
print("Predictor: %s, Rank: %i, Score: %r" % (predictor, rank, score))
# Get the Lakshmanan-like multipass results
print("Multipass. This should have exactly 8 items")
multipass = result.retrieve_multipass()
for predictor in multipass.keys():
rank, score = multipass[predictor]
print("Predictor: %s, Rank: %i, Score: %r" % (predictor, rank, score))
# Iterate over the (context, result) pairs
for i, (cntxt, res) in enumerate(result):
print("Context %i: %r" % (i, cntxt))
print("Result %i: %r" % (i, res))
# ------------------------------------------------------------------------------
# Use the plotting code in examples/plotting.py, found here:
# https://github.com/gelijergensen/PermutationImportance
try:
from plotting import plot_variable_importance
except Exception as e:
print("An error occurred while plotting. You probably don't have matplotlib installed")
print(e)
else:
plot_variable_importance(
result, 'example_singlepass_permutation.png', multipass=False)
plot_variable_importance(
result, 'example_multipass_permutation.png', multipass=True)
Method-Specific¶
-
PermutationImportance.permutation_importance.permutation_importance(scoring_data, scoring_fn, scoring_strategy, variable_names=None, nimportant_vars=None, njobs=1)[source] Performs permutation importance over data given a particular set of functions for scoring and determining optimal variables
Parameters: - scoring_data – a 2-tuple
(inputs, outputs)for scoring in thescoring_fn - scoring_fn – a function to be used for scoring. Should be of the form
(training_data, scoring_data) -> some_value - scoring_strategy – a function to be used for determining optimal
variables. Should be of the form
([some_value]) -> index - variable_names – an optional list for variable names. If not given, will use names of columns of data (if pandas dataframe) or column indices
- nimportant_vars – number of variables to compute multipass importance for. Defaults to all variables
- njobs – an integer for the number of threads to use. If negative, will
use
num_cpus + njobs. Defaults to 1
Returns: PermutationImportance.result.ImportanceResultobject which contains the results for each run- scoring_data – a 2-tuple
Example¶
from sklearn.datasets import load_iris
from sklearn.metrics import accuracy_score
from sklearn.neural_network import MLPClassifier
from PermutationImportance import permutation_importance
# Separate out the last 20% for scoring data
iris = load_iris(return_X_y=False)
inputs = iris.get('data')
outputs = iris.get('target')
predictor_names = iris.get('feature_names')
training_inputs = inputs[:int(0.8 * len(inputs))]
training_outputs = outputs[:int(0.8 * len(outputs))]
scoring_inputs = inputs[int(0.8 * len(inputs)):]
scoring_outputs = outputs[int(0.8 * len(outputs)):]
# Some model we are interested in
model = MLPClassifier(solver='lbfgs')
model.fit(training_inputs, training_outputs)
def score_model(training_data, scoring_data):
"""Custom function to use for scoring. Notice that we are using a global
model here, rather than just reassemble the model each time
:param training_data: should be ignored for permutation importance
:param scoring_data: (scoring_inputs, scoring_outputs)
"""
scoring_ins, scoring_outs = scoring_data
return accuracy_score(scoring_outs, model.predict(scoring_ins))
# Package the data into the right shape
scoring_data = (scoring_inputs, scoring_outputs)
# Use the permutation_importance to compute importances
result = permutation_importance(
scoring_data, score_model, 'min', variable_names=predictor_names)
# Get the Breiman-like singlepass results
print("Singlepass")
singlepass = result.retrieve_singlepass()
for predictor in singlepass.keys():
rank, score = singlepass[predictor]
print("Predictor: %s, Rank: %i, Score: %f" % (predictor, rank, score))
# Get the Lakshmanan-like multipass results
print("Multipass")
multipass = result.retrieve_multipass()
for predictor in multipass.keys():
rank, score = multipass[predictor]
print("Predictor: %s, Rank: %i, Score: %f" % (predictor, rank, score))
# Iterate over the (context, result) pairs
for i, (cntxt, res) in enumerate(result):
print("Context %i: %r" % (i, cntxt))
print("Result %i: %r" % (i, res))
References¶
Sequential Selection¶
Theory¶
Sequential selection actually refers to an entire class of data-based methods. While there are many forms, we presently provide an implementation of the simplest two, sequential forward selection and sequential backward selection. A synopsis of these two methods, as well as several generalizations, can be found in Chapter 9 of Webb (2003). Sequential selection methods determine which predictors are important by evaluating model performance on a dataset where only some of the predictors are present. Predictors which, when present, improve the performance are typically considered important and predictors which, when removed, do not or only slightly degrade the performance are typically considered unimportant. In contrast with permutation importance, sequential selection methods train a new model at every step and are generally much more computationally expensive.
Sequential Forward Selection¶
Sequential forward selection iteratively adds predictors to the set of important predictors by taking the predictor at each step which most improves the performance of the model when added to the set of training predictors. This effectively determines the best 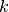 predictors for training a
predictors for training a 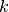 -predictor model. The process is demonstrated in Fig. 1: Sequential forward selection.
-predictor model. The process is demonstrated in Fig. 1: Sequential forward selection.
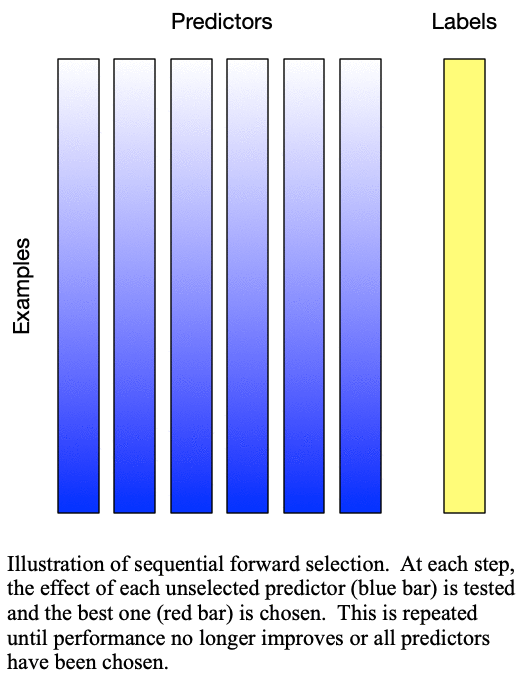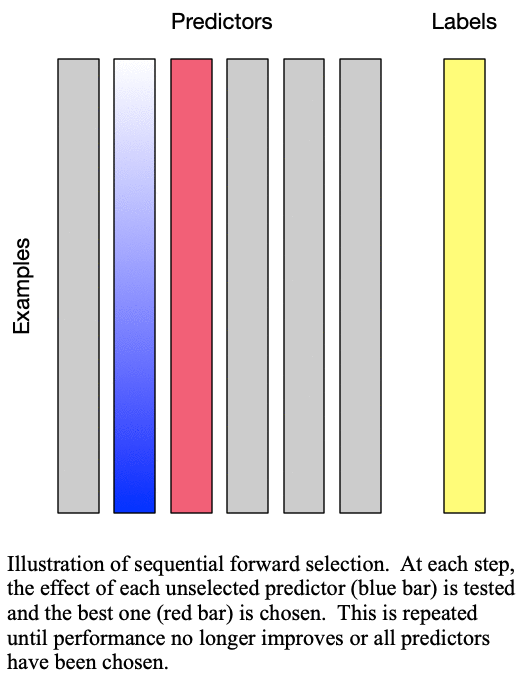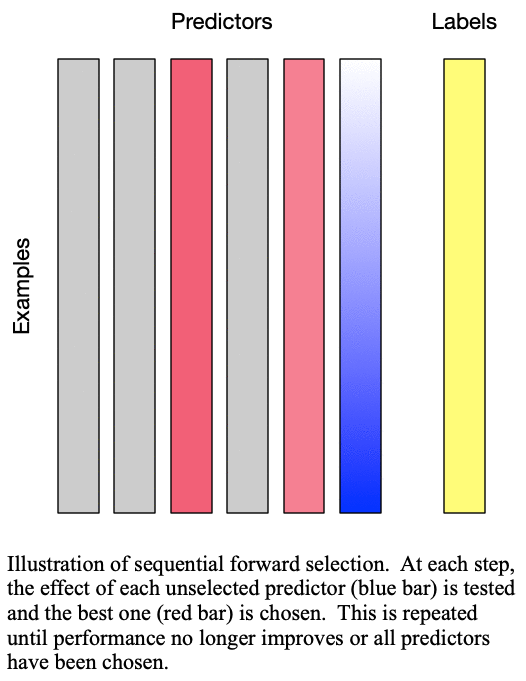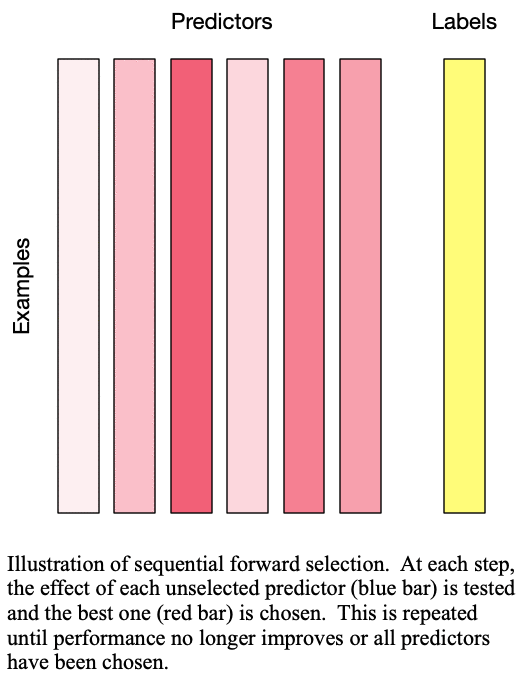
Fig. 1: Sequential forward selection adds the next-best predictor at each step
Sequential Backward Selection¶
Sequential backward selection iteratively removes variables from the set of important variables by taking the predictor at each step which least degrades the performance of the model when removed from the set of training predictors. This effectively determines the  least important predictors. The process is demonstrated in Fig. 2: Sequential backward selection. A word of caution: sequential backward selection can take many times longer than sequential forward selection because it is training many more models with nearly complete sets of predictors. When there are more than 50 predictors, sequential backward selection often becomes computationally infeasible for some models.
least important predictors. The process is demonstrated in Fig. 2: Sequential backward selection. A word of caution: sequential backward selection can take many times longer than sequential forward selection because it is training many more models with nearly complete sets of predictors. When there are more than 50 predictors, sequential backward selection often becomes computationally infeasible for some models.
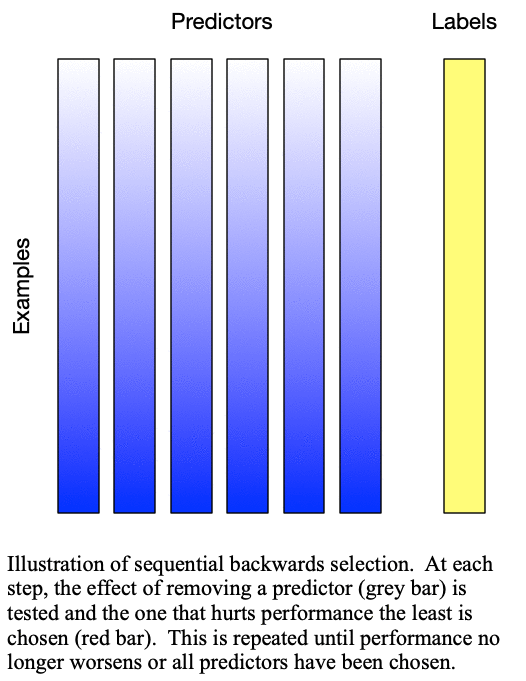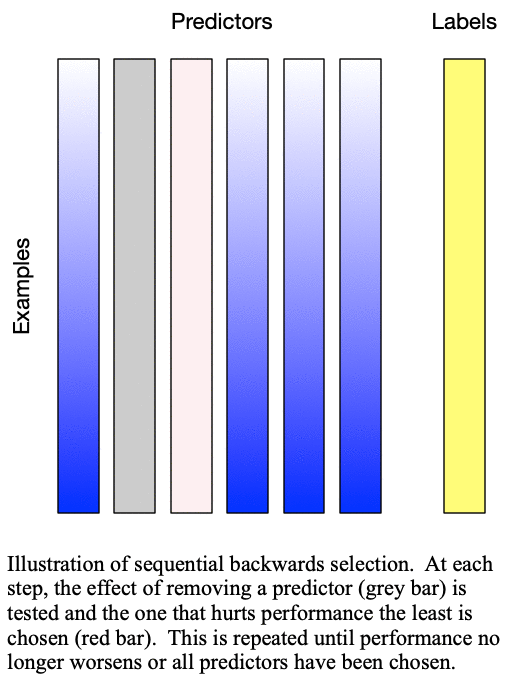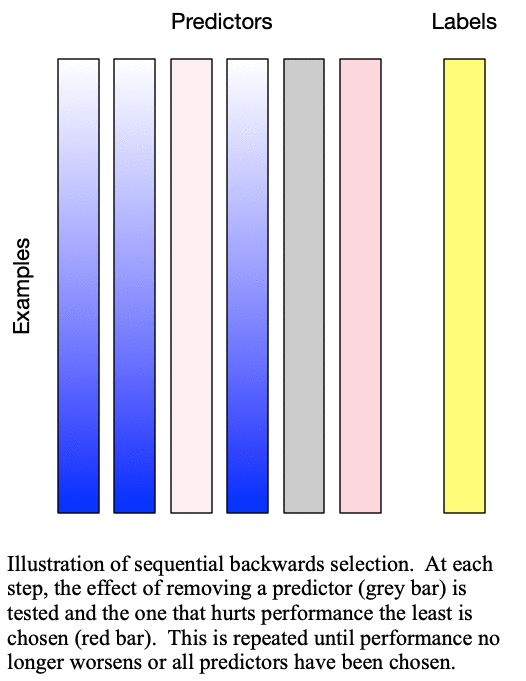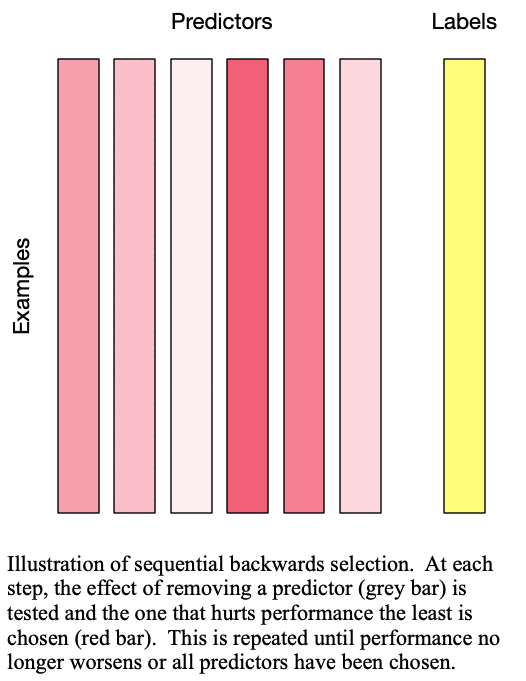
Fig. 2: Sequential backward selection removes the next-worst predictor at each step
Usage¶
As with all methods, we provide all sequential forward selection methods at two different levels of abstraction. For more information on the levels of abstraction and when to use each, please see Levels of Abstraction
Typically, when using a performance metric or skill score with any sequential
selection method, the scoring_strategy should be to maximize the performance.
On the other hand, when using an error or loss function, the scoring_strategy
should be to minimize the error or loss function.
Model-Based¶
-
PermutationImportance.sequential_selection.sklearn_sequential_forward_selection(model, training_data, scoring_data, evaluation_fn, scoring_strategy, variable_names=None, nimportant_vars=None, njobs=1, nbootstrap=None, subsample=1, **kwargs)[source] Performs sequential forward selection for a particular model,
scoring_data,evaluation_fn, and strategy for determining optimal variablesParameters: - model – a sklearn model
- training_data – a 2-tuple
(inputs, outputs)for training in thescoring_fn - scoring_data – a 2-tuple
(inputs, outputs)for scoring in thescoring_fn - evaluation_fn –
a function which takes the deterministic or probabilistic model predictions and scores them against the true values. Must be of the form
(truths, predictions) -> some_valueProbably one of the metrics inPermutationImportance.metricsor sklearn.metrics - scoring_strategy – a function to be used for determining optimal
variables. Should be of the form
([some_value]) -> index - variable_names – an optional list for variable names. If not given, will use names of columns of data (if pandas dataframe) or column indices
- nimportant_vars – number of variables to compute importance for. Defaults to all variables
- njobs – an integer for the number of threads to use. If negative, will
use
num_cpus + njobs. Defaults to 1 - nbootstrap – number of times to perform scoring on each variable. Results over different bootstrap iterations are averaged. Defaults to 1
- subsample – number of elements to sample (with replacement) per bootstrap round. If between 0 and 1, treated as a fraction of the number of total number of events (e.g. 0.5 means half the number of events). If not specified, subsampling will not be used and the entire data will be used (without replacement)
- kwargs – all other kwargs will be passed on to the
evaluation_fn
Returns: PermutationImportance.result.ImportanceResultobject which contains the results for each run
Simple SFS Example¶
from sklearn.datasets import load_iris
from sklearn.metrics import accuracy_score
from sklearn.neural_network import MLPClassifier
from PermutationImportance import sklearn_sequential_forward_selection
# Separate out the last 20% for scoring data
iris = load_iris(return_X_y=False)
inputs = iris.get('data')
outputs = iris.get('target')
predictor_names = iris.get('feature_names')
training_inputs = inputs[:int(0.8 * len(inputs))]
training_outputs = outputs[:int(0.8 * len(outputs))]
scoring_inputs = inputs[int(0.8 * len(inputs)):]
scoring_outputs = outputs[int(0.8 * len(outputs)):]
# Train a quick neural net on the data
model = MLPClassifier(solver='lbfgs')
model.fit(training_inputs, training_outputs)
# Package the data into the right shape
training_data = (training_inputs, training_outputs)
scoring_data = (scoring_inputs, scoring_outputs)
# Use the sklearn_sequential_forward_selection to compute importances
result = sklearn_sequential_forward_selection(
model, training_data, scoring_data, accuracy_score, 'max',
variable_names=predictor_names)
# Get the Breiman-like singlepass results
print("Singlepass")
singlepass = result.retrieve_singlepass()
for predictor in singlepass.keys():
rank, score = singlepass[predictor]
print("Predictor: %s, Rank: %i, Score: %f" % (predictor, rank, score))
# Get the Lakshmanan-like multipass results
print("Multipass")
multipass = result.retrieve_multipass()
for predictor in multipass.keys():
rank, score = multipass[predictor]
print("Predictor: %s, Rank: %i, Score: %f" % (predictor, rank, score))
# Iterate over the (context, result) pairs
for i, (cntxt, res) in enumerate(result):
print("Context %i: %r" % (i, cntxt))
print("Result %i: %r" % (i, res))
Complex SFS Example¶
from sklearn.datasets import load_breast_cancer
from sklearn.ensemble import RandomForestClassifier
from PermutationImportance import sklearn_sequential_forward_selection
from PermutationImportance.metrics import peirce_skill_score
# Separate out the last 20% for scoring data
breast_cancer = load_breast_cancer(return_X_y=False)
inputs = breast_cancer.get('data')
outputs = breast_cancer.get('target')
predictor_names = breast_cancer.get('feature_names')
training_inputs = inputs[:int(0.8 * len(inputs))]
training_outputs = outputs[:int(0.8 * len(outputs))]
scoring_inputs = inputs[int(0.8 * len(inputs)):]
scoring_outputs = outputs[int(0.8 * len(outputs)):]
# Train a quick forest on the data
model = RandomForestClassifier(n_estimators=100, max_depth=4)
model.fit(training_inputs, training_outputs)
# Package the data into the right shape
training_data = (training_inputs, training_outputs)
scoring_data = (scoring_inputs, scoring_outputs)
# ----------- Version to use when only wanting singlepass results --------------
# Use the sklearn_sequential_forward_selection to compute importances
result = sklearn_sequential_forward_selection(
# argmax_of_mean handles bootstrapped metrics
model, training_data, scoring_data, peirce_skill_score, 'argmax_of_mean',
variable_names=predictor_names,
# sample (with replacement) 1*(number of samples) 5 times to compute metric distribution
# nbootstrap should typically be 1000, but this is kept small here for printing purposes
nbootstrap=5, subsample=1,
# only perform for the very top predictor (effectively means only compute singlepass results)
nimportant_vars=1)
# Get the Breiman-like singlepass results
print("Singlepass")
singlepass = result.retrieve_singlepass()
for predictor in singlepass.keys():
rank, score = singlepass[predictor]
print("Predictor: %s, Rank: %i, Score: %r" % (predictor, rank, score))
# Get the Lakshmanan-like multipass results
print("Multipass. This should only have 1 item and be not very useful")
multipass = result.retrieve_multipass()
for predictor in multipass.keys():
rank, score = multipass[predictor]
print("Predictor: %s, Rank: %i, Score: %r" % (predictor, rank, score))
# Iterate over the (context, result) pairs
for i, (cntxt, res) in enumerate(result):
print("Context %i: %r" % (i, cntxt))
print("Result %i: %r" % (i, res))
# ------------------------------------------------------------------------------
# ----------- Version to use when wanting multipass results --------------------
# Use the sklearn_sequential_forward_selection to compute importances
result = sklearn_sequential_forward_selection(
# argmax_of_mean handles bootstrapped metrics
model, training_data, scoring_data, peirce_skill_score, 'argmax_of_mean',
variable_names=predictor_names,
# sample (with replacement) 1*(number of samples) 5 times to compute metric distribution
# nbootstrap should typically be 1000, but this is kept small here for printing purposes
nbootstrap=5, subsample=1,
nimportant_vars=8) # only perform for the top 8 predictors
# Get the Breiman-like singlepass results
print("Singlepass")
singlepass = result.retrieve_singlepass()
for predictor in singlepass.keys():
rank, score = singlepass[predictor]
print("Predictor: %s, Rank: %i, Score: %r" % (predictor, rank, score))
# Get the Lakshmanan-like multipass results
print("Multipass. This should have exactly 8 items")
multipass = result.retrieve_multipass()
for predictor in multipass.keys():
rank, score = multipass[predictor]
print("Predictor: %s, Rank: %i, Score: %r" % (predictor, rank, score))
# Iterate over the (context, result) pairs
for i, (cntxt, res) in enumerate(result):
print("Context %i: %r" % (i, cntxt))
print("Result %i: %r" % (i, res))
# ------------------------------------------------------------------------------
# Use the plotting code in examples/plotting.py, found here:
# https://github.com/gelijergensen/PermutationImportance
try:
from plotting import plot_variable_importance
except Exception as e:
print("An error occurred while plotting. You probably don't have matplotlib installed")
print(e)
else:
plot_variable_importance(
result, 'example_sequential_forward_selection.png')
-
PermutationImportance.sequential_selection.sklearn_sequential_backward_selection(model, training_data, scoring_data, evaluation_fn, scoring_strategy, variable_names=None, nimportant_vars=None, njobs=1, nbootstrap=None, subsample=1, **kwargs)[source] Performs sequential backward selection for a particular model,
scoring_data,evaluation_fn, and strategy for determining optimal variablesParameters: - model – a sklearn model
- training_data – a 2-tuple
(inputs, outputs)for training in thescoring_fn - scoring_data – a 2-tuple
(inputs, outputs)for scoring in thescoring_fn - evaluation_fn –
a function which takes the deterministic or probabilistic model predictions and scores them against the true values. Must be of the form
(truths, predictions) -> some_valueProbably one of the metrics inPermutationImportance.metricsor sklearn.metrics - scoring_strategy – a function to be used for determining optimal
variables. Should be of the form
([some_value]) -> index - variable_names – an optional list for variable names. If not given, will use names of columns of data (if pandas dataframe) or column indices
- nimportant_vars – number of variables to compute importance for. Defaults to all variables
- njobs – an integer for the number of threads to use. If negative, will
use
num_cpus + njobs. Defaults to 1 - nbootstrap – number of times to perform scoring on each variable. Results over different bootstrap iterations are averaged. Defaults to 1
- subsample – number of elements to sample (with replacement) per bootstrap round. If between 0 and 1, treated as a fraction of the number of total number of events (e.g. 0.5 means half the number of events). If not specified, subsampling will not be used and the entire data will be used (without replacement)
- kwargs – all other kwargs will be passed on to the
evaluation_fn
Returns: PermutationImportance.result.ImportanceResultobject which contains the results for each run
Simple SBS Example¶
from sklearn.datasets import load_iris
from sklearn.metrics import accuracy_score
from sklearn.neural_network import MLPClassifier
from PermutationImportance import sklearn_sequential_backward_selection
# Separate out the last 20% for scoring data
iris = load_iris(return_X_y=False)
inputs = iris.get('data')
outputs = iris.get('target')
predictor_names = iris.get('feature_names')
training_inputs = inputs[:int(0.8 * len(inputs))]
training_outputs = outputs[:int(0.8 * len(outputs))]
scoring_inputs = inputs[int(0.8 * len(inputs)):]
scoring_outputs = outputs[int(0.8 * len(outputs)):]
# Train a quick neural net on the data
model = MLPClassifier(solver='lbfgs')
model.fit(training_inputs, training_outputs)
# Package the data into the right shape
training_data = (training_inputs, training_outputs)
scoring_data = (scoring_inputs, scoring_outputs)
# Use the sklearn_sequential_backward_selection to compute importances
result = sklearn_sequential_backward_selection(
model, training_data, scoring_data, accuracy_score, 'max',
variable_names=predictor_names)
# Get the Breiman-like singlepass results
print("Singlepass")
singlepass = result.retrieve_singlepass()
for predictor in singlepass.keys():
rank, score = singlepass[predictor]
print("Predictor: %s, Rank: %i, Score: %f" % (predictor, rank, score))
# Get the Lakshmanan-like multipass results
print("Multipass")
multipass = result.retrieve_multipass()
for predictor in multipass.keys():
rank, score = multipass[predictor]
print("Predictor: %s, Rank: %i, Score: %f" % (predictor, rank, score))
# Iterate over the (context, result) pairs
for i, (cntxt, res) in enumerate(result):
print("Context %i: %r" % (i, cntxt))
print("Result %i: %r" % (i, res))
Complex SBS Example¶
from sklearn.datasets import load_breast_cancer
from sklearn.ensemble import RandomForestClassifier
from PermutationImportance import sklearn_sequential_backward_selection
from PermutationImportance.metrics import peirce_skill_score
# Separate out the last 20% for scoring data
breast_cancer = load_breast_cancer(return_X_y=False)
inputs = breast_cancer.get('data')
outputs = breast_cancer.get('target')
predictor_names = breast_cancer.get('feature_names')
training_inputs = inputs[:int(0.8 * len(inputs))]
training_outputs = outputs[:int(0.8 * len(outputs))]
scoring_inputs = inputs[int(0.8 * len(inputs)):]
scoring_outputs = outputs[int(0.8 * len(outputs)):]
# Train a quick forest on the data
model = RandomForestClassifier(n_estimators=100, max_depth=4)
model.fit(training_inputs, training_outputs)
# Package the data into the right shape
training_data = (training_inputs, training_outputs)
scoring_data = (scoring_inputs, scoring_outputs)
# ----------- Version to use when only wanting singlepass results --------------
# Use the sklearn_sequential_backward_selection to compute importances
result = sklearn_sequential_backward_selection(
# argmax_of_mean handles bootstrapped metrics
model, training_data, scoring_data, peirce_skill_score, 'argmax_of_mean',
variable_names=predictor_names,
# sample (with replacement) 1*(number of samples) 5 times to compute metric distribution
# nbootstrap should typically be 1000, but this is kept small here for printing purposes
nbootstrap=5, subsample=1,
# only perform for the very top predictor (effectively means only compute singlepass results)
nimportant_vars=1)
# Get the Breiman-like singlepass results
print("Singlepass")
singlepass = result.retrieve_singlepass()
for predictor in singlepass.keys():
rank, score = singlepass[predictor]
print("Predictor: %s, Rank: %i, Score: %r" % (predictor, rank, score))
# Get the Lakshmanan-like multipass results
print("Multipass. This should only have 1 item and be not very useful")
multipass = result.retrieve_multipass()
for predictor in multipass.keys():
rank, score = multipass[predictor]
print("Predictor: %s, Rank: %i, Score: %r" % (predictor, rank, score))
# Iterate over the (context, result) pairs
for i, (cntxt, res) in enumerate(result):
print("Context %i: %r" % (i, cntxt))
print("Result %i: %r" % (i, res))
# ------------------------------------------------------------------------------
# ----------- Version to use when wanting multipass results --------------------
# Use the sklearn_sequential_backward_selection to compute importances
result = sklearn_sequential_backward_selection(
# argmax_of_mean handles bootstrapped metrics
model, training_data, scoring_data, peirce_skill_score, 'argmax_of_mean',
variable_names=predictor_names,
# sample (with replacement) 1*(number of samples) 5 times to compute metric distribution
# nbootstrap should typically be 1000, but this is kept small here for printing purposes
nbootstrap=5, subsample=1,
nimportant_vars=8) # only perform for the top 8 predictors
# Get the Breiman-like singlepass results
print("Singlepass")
singlepass = result.retrieve_singlepass()
for predictor in singlepass.keys():
rank, score = singlepass[predictor]
print("Predictor: %s, Rank: %i, Score: %r" % (predictor, rank, score))
# Get the Lakshmanan-like multipass results
print("Multipass. This should have exactly 8 items")
multipass = result.retrieve_multipass()
for predictor in multipass.keys():
rank, score = multipass[predictor]
print("Predictor: %s, Rank: %i, Score: %r" % (predictor, rank, score))
# Iterate over the (context, result) pairs
for i, (cntxt, res) in enumerate(result):
print("Context %i: %r" % (i, cntxt))
print("Result %i: %r" % (i, res))
# ------------------------------------------------------------------------------
# Use the plotting code in examples/plotting.py, found here:
# https://github.com/gelijergensen/PermutationImportance
try:
from plotting import plot_variable_importance
except Exception as e:
print("An error occurred while plotting. You probably don't have matplotlib installed")
print(e)
else:
plot_variable_importance(
result, 'example_sequential_backward_selection.png')
Method-Specific¶
-
PermutationImportance.sequential_selection.sequential_forward_selection(training_data, scoring_data, scoring_fn, scoring_strategy, variable_names=None, nimportant_vars=None, njobs=1)[source] Performs sequential forward selection over data given a particular set of functions for scoring and determining optimal variables
Parameters: - training_data – a 2-tuple
(inputs, outputs)for training in thescoring_fn - scoring_data – a 2-tuple
(inputs, outputs)for scoring in thescoring_fn - scoring_fn – a function to be used for scoring. Should be of the form
(training_data, scoring_data) -> some_value - scoring_strategy – a function to be used for determining optimal
variables. Should be of the form
([some_value]) -> index - variable_names – an optional list for variable names. If not given, will use names of columns of data (if pandas dataframe) or column indices
- nimportant_vars – number of variables to compute importance for. Defaults to all variables
- njobs – an integer for the number of threads to use. If negative, will
use
num_cpus + njobs. Defaults to 1
Returns: PermutationImportance.result.ImportanceResultobject which contains the results for each run- training_data – a 2-tuple
SFS Example¶
from sklearn.datasets import load_iris
from sklearn.metrics import accuracy_score
from sklearn.neural_network import MLPClassifier
from PermutationImportance import sequential_forward_selection
# Separate out the last 20% for scoring data
iris = load_iris(return_X_y=False)
inputs = iris.get('data')
outputs = iris.get('target')
predictor_names = iris.get('feature_names')
training_inputs = inputs[:int(0.8 * len(inputs))]
training_outputs = outputs[:int(0.8 * len(outputs))]
scoring_inputs = inputs[int(0.8 * len(inputs)):]
scoring_outputs = outputs[int(0.8 * len(outputs)):]
def score_model(training_data, scoring_data):
"""Custom function to use for scoring. Notice that because this is
sequential selection, we need to retrain the model each time
:param training_data: (training_inputs, training_outputs)
:param scoring_data: (scoring_inputs, scoring_outputs)
"""
training_ins, training_outs = training_data
scoring_ins, scoring_outs = scoring_data
# Some model we are interested in
model = MLPClassifier(solver='lbfgs')
model.fit(training_ins, training_outs)
return accuracy_score(scoring_outs, model.predict(scoring_ins))
# Package the data into the right shape
training_data = (training_inputs, training_outputs)
scoring_data = (scoring_inputs, scoring_outputs)
# Use the sequential_forward_selection to compute importances
result = sequential_forward_selection(
training_data, scoring_data, score_model, 'max',
variable_names=predictor_names)
# Get the Breiman-like singlepass results
print("Singlepass")
singlepass = result.retrieve_singlepass()
for predictor in singlepass.keys():
rank, score = singlepass[predictor]
print("Predictor: %s, Rank: %i, Score: %f" % (predictor, rank, score))
# Get the Lakshmanan-like multipass results
print("Multipass")
multipass = result.retrieve_multipass()
for predictor in multipass.keys():
rank, score = multipass[predictor]
print("Predictor: %s, Rank: %i, Score: %f" % (predictor, rank, score))
# Iterate over the (context, result) pairs
for i, (cntxt, res) in enumerate(result):
print("Context %i: %r" % (i, cntxt))
print("Result %i: %r" % (i, res))
-
PermutationImportance.sequential_selection.sequential_backward_selection(training_data, scoring_data, scoring_fn, scoring_strategy, variable_names=None, nimportant_vars=None, njobs=1)[source] Performs sequential backward selection over data given a particular set of functions for scoring and determining optimal variables
Parameters: - training_data – a 2-tuple
(inputs, outputs)for training in thescoring_fn - scoring_data – a 2-tuple
(inputs, outputs)for scoring in thescoring_fn - scoring_fn – a function to be used for scoring. Should be of the form
(training_data, scoring_data) -> some_value - scoring_strategy – a function to be used for determining optimal
variables. Should be of the form
([some_value]) -> index - variable_names – an optional list for variable names. If not given, will use names of columns of data (if pandas dataframe) or column indices
- nimportant_vars – number of variables to compute importance for. Defaults to all variables
- njobs – an integer for the number of threads to use. If negative, will
use
num_cpus + njobs. Defaults to 1
Returns: PermutationImportance.result.ImportanceResultobject which contains the results for each run- training_data – a 2-tuple
SBS Example¶
from sklearn.datasets import load_iris
from sklearn.metrics import accuracy_score
from sklearn.neural_network import MLPClassifier
from PermutationImportance import sequential_backward_selection
# Separate out the last 20% for scoring data
iris = load_iris(return_X_y=False)
inputs = iris.get('data')
outputs = iris.get('target')
predictor_names = iris.get('feature_names')
training_inputs = inputs[:int(0.8 * len(inputs))]
training_outputs = outputs[:int(0.8 * len(outputs))]
scoring_inputs = inputs[int(0.8 * len(inputs)):]
scoring_outputs = outputs[int(0.8 * len(outputs)):]
def score_model(training_data, scoring_data):
"""Custom function to use for scoring. Notice that because this is
sequential selection, we need to retrain the model each time
:param training_data: (training_inputs, training_outputs)
:param scoring_data: (scoring_inputs, scoring_outputs)
"""
training_ins, training_outs = training_data
scoring_ins, scoring_outs = scoring_data
# Some model we are interested in
model = MLPClassifier(solver='lbfgs')
model.fit(training_ins, training_outs)
return accuracy_score(scoring_outs, model.predict(scoring_ins))
# Package the data into the right shape
training_data = (training_inputs, training_outputs)
scoring_data = (scoring_inputs, scoring_outputs)
# Use the sequential_backward_selection to compute importances
result = sequential_backward_selection(
training_data, scoring_data, score_model, 'max',
variable_names=predictor_names)
# Get the Breiman-like singlepass results
print("Singlepass")
singlepass = result.retrieve_singlepass()
for predictor in singlepass.keys():
rank, score = singlepass[predictor]
print("Predictor: %s, Rank: %i, Score: %f" % (predictor, rank, score))
# Get the Lakshmanan-like multipass results
print("Multipass")
multipass = result.retrieve_multipass()
for predictor in multipass.keys():
rank, score = multipass[predictor]
print("Predictor: %s, Rank: %i, Score: %f" % (predictor, rank, score))
# Iterate over the (context, result) pairs
for i, (cntxt, res) in enumerate(result):
print("Context %i: %r" % (i, cntxt))
print("Result %i: %r" % (i, res))
Custom Methods¶
While we provide a number of data-based methods out of the box, you may find that you wish to implement a data-based predictor importance method which we have not provided. For convenience, we provide tools that may assist in the process of implementing those methods.
Firstly, we provide the function abstract_variable_importance, which encapsulates the general process of performing a data-based predictor importance method and additionally provides automatic hooks into both the single- and multi-process backends. So long as the method which you wish to implement follows the general structure of “scoring” given (training_data, scoring_data) tuples to evaluate importance for each predictor in succession, you should be able to use the abstract_variable_importance function directly by only providing a valid selection_strategy. For more on this process, so below. Even if your desired method does not match this pattern, you may still find utility in the two backends in PermutationImportance.abstract_runner.
Abstract Variable Importance¶
The abstract_variable_importance function handles the generalized process for computing predictor importance. The algorithm itself consists of a double-for loop, the first of which loops once for each of the predictors, the second of which loops over the list of triples (predictor, training_data, scoring_data) returned by the given selection_strategy. This allows for the majority of the implementation details to be left to the selection_strategy.
-
PermutationImportance.abstract_runner.abstract_variable_importance(training_data, scoring_data, scoring_fn, scoring_strategy, selection_strategy, variable_names=None, nimportant_vars=None, method=None, njobs=1)[source] Performs an abstract variable importance over data given a particular set of functions for scoring, determining optimal variables, and selecting data
Parameters: - training_data – a 2-tuple
(inputs, outputs)for training in thescoring_fn - scoring_data – a 2-tuple
(inputs, outputs)for scoring in thescoring_fn - scoring_fn – a function to be used for scoring. Should be of the form
(training_data, scoring_data) -> some_value - scoring_strategy – a function to be used for determining optimal
variables. Should be of the form
([some_value]) -> index - variable_names – an optional list for variable names. If not given, will use names of columns of data (if pandas dataframe) or column indices
- nimportant_vars – number of variables to compute importance for. Defaults to all variables
- method – a string for the name of the method used. Defaults to the
name of the
selection_strategyif not given - njobs – an integer for the number of threads to use. If negative, will
use
num_cpus + njobs. Defaults to 1
Returns: PermutationImportance.result.ImportanceResultobject which contains the results for each run- training_data – a 2-tuple
Selection Strategy¶
The selection strategy is the most important part of a predictor importance method, as it essentially defines the method. Here, a SelectionStrategy is an object which is initialized with the original training_data and scoring_data datasets passed to the predictor importance method, the total number of variables, and the current variables which are considered important. It must act as a generator which yields tuples of (variable, training_data_subset, scoring_data_subset). This can be thought of as yielding the information to test the importance of this variable by using the training_data_subset and scoring_data_subset.
For convenience, we provide the base SelectionStrategy object, which should be extended to make a new method. Each object should have a static name property (for diagnostics) and should override the generate_all_datasets or generate_datasets method. As many methods test precisely the predictors which are not yet considered important, the default implementation of generate_all_datasets calls generate_datasets once for each currently unimportant predictor. Please see the implementation of the base SelectionStrategy object, as well as the other classes in PermutationImportance.selection_strategies for more details.
-
class
PermutationImportance.selection_strategies.SelectionStrategy(training_data, scoring_data, num_vars, important_vars)[source] The base
SelectionStrategyonly provides the tools for storing the data and other important information as well as the convenience method for iterating over the selection strategies triples lazily.-
__init__(training_data, scoring_data, num_vars, important_vars)[source] Initializes the object by storing the data and keeping track of other important information
Parameters: - training_data – (training_inputs, training_outputs)
- scoring_data – (scoring_inputs, scoring_outputs)
- num_vars – integer for the total number of variables
- important_vars – a list of the indices of variables which are already considered important
-
generate_all_datasets()[source] By default, loops over all variables not yet considered important
-
generate_datasets(important_variables)[source] Generator which returns triples (variable, training_data, scoring_data)
-
Example of Custom Variable Importance¶
"""An example of several different custom components that PermutationImportance
allows. Here, we are attempting to look at the predictors which are impacting
the forecasting bias of the model. To do this, we first construct a custom
metric ``bias_score``, and also construct an optimization strategy which selects
the index of the predictor which induces the least bias in the model
``argmin_of_ratio_from_unity``. Additionally, rather than using a typical method
for evaluating this (like permutation importance), we develop our own custom
method, "zero-filled importance", which operates like permutation importance,
but rather than permuting the values of a predictor to destroy the relationship
between the predictor and the target, it simply sets all of the values of the
predictor to 0 (which could have some interesting, undesired side-effects). This
is done by constructing a custom selection strategy
``ZeroFilledSelectionStrategy`` and using this to build both the method-specific
(``zero_filled_importance``) and model-based
(``sklearn_zero_filled_importance``) versions of the predictor importance
method.
As a side note, notice below that we leverage the utilities of
PermutationImportance.sklearn_api to help build the model-based version in a way
which also allows us to even do bootstrapping!
"""
from sklearn.ensemble import RandomForestClassifier
from sklearn.datasets import load_breast_cancer
import numpy as np
from PermutationImportance.abstract_runner import abstract_variable_importance
from PermutationImportance.metrics import _get_contingency_table
from PermutationImportance.selection_strategies import SelectionStrategy
from PermutationImportance.sklearn_api import score_trained_sklearn_model, score_trained_sklearn_model_with_probabilities
from PermutationImportance.utils import get_data_subset, make_data_from_columns
# Example of a custom metric / evaluation_fn
def bias_score(truths, predictions, classes=None):
"""Determines the Forecast Bias of a model, returning a scalar. See
`here <http://www.cawcr.gov.au/projects/verification/#Methods_for_dichotomous_forecasts>`_
for more details on the bias score.
To handle multi-class predictions, this takes the AVERAGE bias score for
each of the classes independently
:param truths: The true labels of these data
:param predictions: The predictions of the model
:param classes: an ordered set for the label possibilities. If not given,
will be deduced from the truth values
:returns: a single value for the gerrity score
"""
table = _get_contingency_table(truths, predictions, classes)
biases = np.zeros((len(table), ))
for i in range(len(biases)):
biases[i] = np.sum(table[i, :], dtype='float32') / \
(np.sum(table[:, i], dtype='float32') +
1e-16) # epsilon for numerical stability
return np.average(biases)
def _ratio_from_unity(score):
"""Returns the smaller of (score, 1/score). This can be thought of as a
score in [0, 1], where 1 is the best and 0 is the worst
:param score: either a single value or an array of values, in which case
the mean is taken first
:returns: a single scalar in [0, 1], where 1 is best"""
mean_score = np.average(score)
if mean_score > 1:
return 1.0 / float(mean_score)
else:
return float(mean_score)
# Example of a custom optimization strategy
def argmin_of_ratio_from_unity(scores):
"""Returns the argmin of each of the "ratios from unity". This has the
effect of returning the index of the predictor which caused the worst bias
NOTE: This could have also been done with
:class:`PermutationImportance.scoring_strategies.indexer_of_converter```(np.armin, _ratio_from_unity)``
"""
return np.argmin([_ratio_from_unity(score) for score in scores])
# Example of a custom selection strategy
class ZeroFilledSelectionStrategy(SelectionStrategy):
""""Zero-Filled Importance" is a made-up predictor importance method which
tests all predictors which are not yet considered importance by setting all
of the values of that column to be zero. This destroys the information
present in the column much in the same way as Permutation Importance, but
may have weird side-effects because zero is not necessarily a neutral value
(e.g. Temperature in kelvins). The shape of the training data will remain
constant, but many columns may contain only 0's."""
name = "Zero Filled Importance"
def __init__(self, training_data, scoring_data, num_vars, important_vars):
"""Initializes the object by storing the data and keeping track of other
important information
:param training_data: (training_inputs, training_outputs)
:param scoring_data: (scoring_inputs, scoring_outputs)
:param num_vars: integer for the total number of variables
:param important_vars: a list of the indices of variables which are
already considered important
"""
super(ZeroFilledSelectionStrategy, self).__init__(
training_data, scoring_data, num_vars, important_vars)
# Also initialize the zero data
scoring_inputs, __ = self.scoring_data
self.zero_scoring_inputs = np.zeros(scoring_inputs.shape)
def generate_datasets(self, important_variables):
"""Check each of the non-important variables. Dataset has columns which
are important shuffled. Notice that although we could modify the
training data as well, we are going to assume that this behaves like
Permutation Importance, in which case the training data will always be
empty
:returns: (training_data, scoring_data)
"""
scoring_inputs, scoring_outputs = self.scoring_data
complete_scoring_inputs = make_data_from_columns(
[get_data_subset(self.zero_scoring_inputs if i in important_variables else scoring_inputs, None, [i]) for i in range(self.num_vars)])
return self.training_data, (complete_scoring_inputs, scoring_outputs)
# Example of the Method-Specific custom predictor importance
def zero_filled_importance(scoring_data, scoring_fn, scoring_strategy, variable_names=None, nimportant_vars=None, njobs=1):
"""Performs "zero-filled importance" over data given a particular
set of functions for scoring and determining optimal variables
:param scoring_data: a 2-tuple ``(inputs, outputs)`` for scoring in the
``scoring_fn``
:param scoring_fn: a function to be used for scoring. Should be of the form
``(training_data, scoring_data) -> some_value``
:param scoring_strategy: a function to be used for determining optimal
variables. Should be of the form ``([some_value]) -> index``
:param variable_names: an optional list for variable names. If not given,
will use names of columns of data (if pandas dataframe) or column
indices
:param nimportant_vars: number of variables to compute multipass importance
for. Defaults to all variables
:param njobs: an integer for the number of threads to use. If negative, will
use ``num_cpus + njobs``. Defaults to 1
:returns: :class:`PermutationImportance.result.ImportanceResult` object
which contains the results for each run
"""
# We don't need the training data, so pass empty arrays to the abstract runner
return abstract_variable_importance((np.array([]), np.array([])), scoring_data, scoring_fn, scoring_strategy, ZeroFilledSelectionStrategy, variable_names=variable_names, nimportant_vars=nimportant_vars, njobs=njobs)
# Example of a Model-Based custom predictor importance
def sklearn_zero_filled_importance(model, scoring_data, evaluation_fn, scoring_strategy, variable_names=None, nimportant_vars=None, njobs=1, nbootstrap=1, subsample=1, **kwargs):
"""Performs "zero-filled importance" for a particular model,
``scoring_data``, ``evaluation_fn``, and strategy for determining optimal
variables
:param model: a trained sklearn model
:param scoring_data: a 2-tuple ``(inputs, outputs)`` for scoring in the
``scoring_fn``
:param evaluation_fn: a function which takes the deterministic or
probabilistic model predictions and scores them against the true
values. Must be of the form ``(truths, predictions) -> some_value``
Probably one of the metrics in
:mod:`PermutationImportance.metrics` or
`sklearn.metrics <https://scikit-learn.org/stable/modules/classes.html#module-sklearn.metrics>`_
:param scoring_strategy: a function to be used for determining optimal
variables. Should be of the form ``([some_value]) -> index``
:param variable_names: an optional list for variable names. If not given,
will use names of columns of data (if pandas dataframe) or column
indices
:param nimportant_vars: number of variables to compute multipass importance
for. Defaults to all variables
:param njobs: an integer for the number of threads to use. If negative, will
use ``num_cpus + njobs``. Defaults to 1
:param nbootstrap: number of times to perform scoring on each variable.
Results over different bootstrap iterations are averaged. Defaults to 1
:param subsample: number of elements to sample (with replacement) per
bootstrap round. If between 0 and 1, treated as a fraction of the number
of total number of events (e.g. 0.5 means half the number of events).
If not specified, subsampling will not be used and the entire data will
be used (without replacement)
:param kwargs: all other kwargs will be passed on to the ``evaluation_fn``
:returns: :class:`PermutationImportance.result.ImportanceResult` object
which contains the results for each run
"""
# Check if the data is probabilistic
if len(scoring_data[1].shape) > 1 and scoring_data[1].shape[1] > 1:
# Take advantage of the tools in PermutationImportance.sklearn_api to
# build a probabilistic scoring function from the evaluation function
scoring_fn = score_trained_sklearn_model_with_probabilities(
model, evaluation_fn, nbootstrap=nbootstrap, subsample=subsample, **kwargs)
else:
# Take advantage of the tools in PermutationImportance.sklearn_api to
# build a deterministic scoring function from the evaluation function
scoring_fn = score_trained_sklearn_model(
model, evaluation_fn, nbootstrap=nbootstrap, subsample=subsample, **kwargs)
return zero_filled_importance(scoring_data, scoring_fn, scoring_strategy, variable_names=variable_names, nimportant_vars=nimportant_vars, njobs=njobs)
# ----------------- Example Usage of Custom Method -----------------------------
"""
Here, our goal is to try and determine which predictors most drastically impact
the bias of a model. Notice that the "_ratio_from_unity" above basically acts
as a way to convert the bias to a more traditional score. Here, we also use a
custom predictor importance method, "Zero-Filled Importance"
"""
# Separate out the last 20% for scoring data
breast_cancer = load_breast_cancer(return_X_y=False)
inputs = breast_cancer.get('data')
outputs = breast_cancer.get('target')
predictor_names = breast_cancer.get('feature_names')
training_inputs = inputs[:int(0.8 * len(inputs))]
training_outputs = outputs[:int(0.8 * len(outputs))]
scoring_inputs = inputs[int(0.8 * len(inputs)):]
scoring_outputs = outputs[int(0.8 * len(outputs)):]
# Train a quick forest on the data
model = RandomForestClassifier(n_estimators=100, max_depth=4)
model.fit(training_inputs, training_outputs)
# Package the data into the right shape
scoring_data = (scoring_inputs, scoring_outputs)
# Use the sklearn_zero_filled_importance to compute importances
result = sklearn_zero_filled_importance(
model, scoring_data, bias_score, argmin_of_ratio_from_unity,
variable_names=predictor_names,
# Notice that we can use bootstrapping here thanks to the
# PermutationImportance.sklearn_api tools for constructing a score function
nbootstrap=5, subsample=1, # nboostrap=1000 would be better
nimportant_vars=None) # perform for all predictors
# Get the Breiman-like singlepass results
print("Singlepass")
singlepass = result.retrieve_singlepass()
for predictor in singlepass.keys():
rank, score = singlepass[predictor]
print("Predictor: %s, Rank: %i, Score: %r" % (predictor, rank, score))
# Get the Lakshmanan-like multipass results
print("Multipass. This should have exactly 8 items")
multipass = result.retrieve_multipass()
for predictor in multipass.keys():
rank, score = multipass[predictor]
print("Predictor: %s, Rank: %i, Score: %r" % (predictor, rank, score))
# Iterate over the (context, result) pairs
for i, (cntxt, res) in enumerate(result):
print("Context %i: %r" % (i, cntxt))
print("Result %i: %r" % (i, res))
# ------------------------------------------------------------------------------