Permutation Importance¶
Theory¶
Permutation importance, the namesake for this package, determines the predictors which are important by comparing the performance of a model on a dataset where some of the predictors are individually permuted to performance on the original dataset without any permutation. The permutation of the individual predictor in this manner effectively breaks the relationship between the input predictor and the target variable. The predictor which, when permuted, results in the worst performance is typically taken as the most important variable. Permutation importance has the distinct advantage of not needing to retrain the model each time.
This method was originally designed for random forests by Breiman (2001), but can be used by any model. The original version of the algorithm was 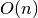 , but this was later revised by Lakshmanan (2015) to be more robust to correlated predictors and is
, but this was later revised by Lakshmanan (2015) to be more robust to correlated predictors and is 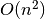 . The difference between these two methods is detailed in Fig. 1: Singlepass permutation importance and Fig. 2: Multipass permutation importance. While Breiman’s method only permutes each predictor once independently, Lakshmanan’s method iteratively adds one predictor to the set of predictors which are permuted at each iteration. Because Lakshmanan’s method can be viewed as successively applying Breiman’s method to determine the next-most important predictor, we typically refer to Breiman’s method as “singlepass” and Lakshmanan’s method as “multipass”.
. The difference between these two methods is detailed in Fig. 1: Singlepass permutation importance and Fig. 2: Multipass permutation importance. While Breiman’s method only permutes each predictor once independently, Lakshmanan’s method iteratively adds one predictor to the set of predictors which are permuted at each iteration. Because Lakshmanan’s method can be viewed as successively applying Breiman’s method to determine the next-most important predictor, we typically refer to Breiman’s method as “singlepass” and Lakshmanan’s method as “multipass”.
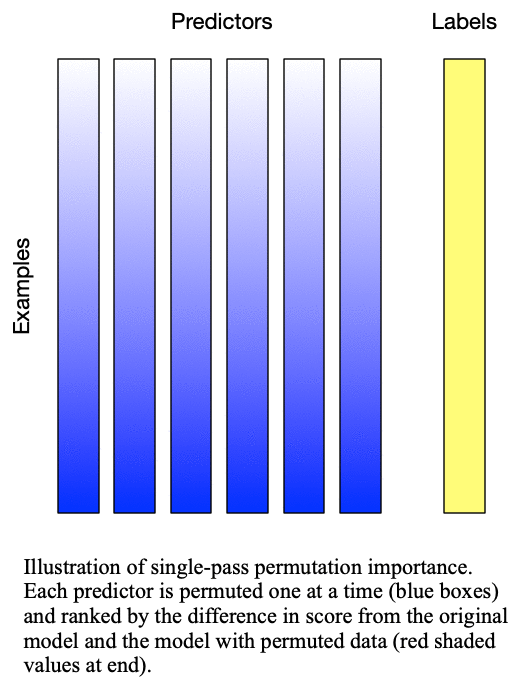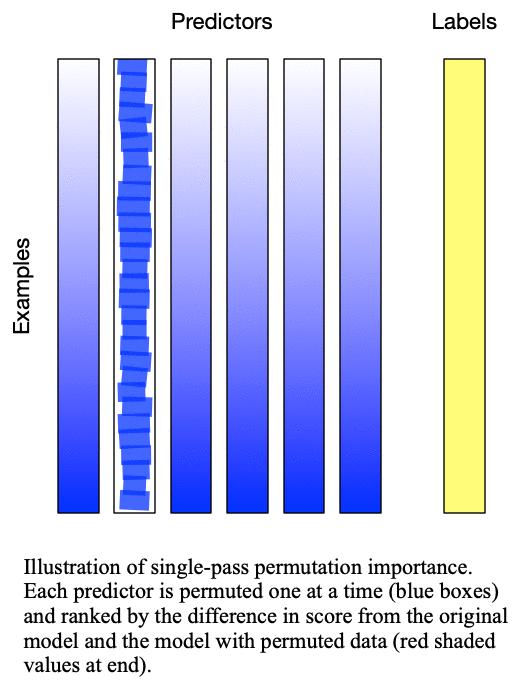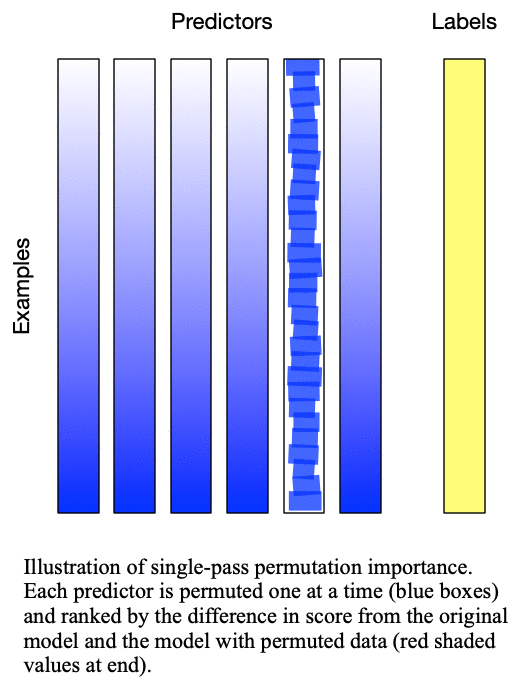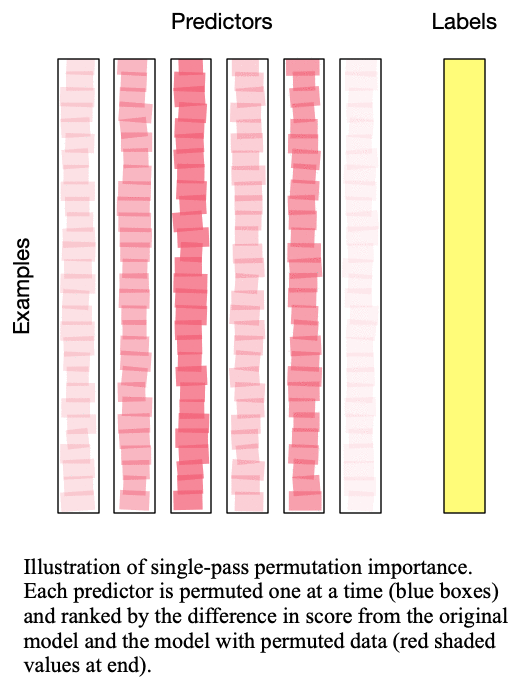
Fig. 1: Singlepass permutation importance evaluates each predictor independently by permuting only the values of that predictor
Fig. 2: Multipass permutation importance performs singlepass permutation importance as many times as there as predictors to iteratively determine the next-most important predictor
Usage¶
As with all methods, we provide the permutation importance method at two different levels of abstraction. For more information on the levels of abstraction and when to use each, please see Levels of Abstraction.
Singlepass permutation importance is computed as a byproduct of the generalized method. To compute singlepass permutation importance only, set nimportant_vars=1, which will only perform the multipass method for precisely one pass.
Typically, when using a performance metric or skill score with permutation
importance, the scoring_strategy should be to minimize the performance. On the
other hand, when using an error or loss function, the scoring_strategy should
be to maximize the error or loss function.
Model-Based¶
-
PermutationImportance.permutation_importance.sklearn_permutation_importance(model, scoring_data, evaluation_fn, scoring_strategy, variable_names=None, nimportant_vars=None, njobs=1, nbootstrap=1, subsample=1, **kwargs)[source] Performs permutation importance for a particular model,
scoring_data,evaluation_fn, and strategy for determining optimal variablesParameters: - model – a trained sklearn model
- scoring_data – a 2-tuple
(inputs, outputs)for scoring in thescoring_fn - evaluation_fn – a function which takes the deterministic or
probabilistic model predictions and scores them against the true
values. Must be of the form
(truths, predictions) -> some_valueProbably one of the metrics inPermutationImportance.metricsor sklearn.metrics - scoring_strategy – a function to be used for determining optimal
variables. Should be of the form
([some_value]) -> index - variable_names – an optional list for variable names. If not given, will use names of columns of data (if pandas dataframe) or column indices
- nimportant_vars – number of variables to compute multipass importance for. Defaults to all variables
- njobs – an integer for the number of threads to use. If negative, will
use
num_cpus + njobs. Defaults to 1 - nbootstrap – number of times to perform scoring on each variable. Results over different bootstrap iterations are averaged. Defaults to 1
- subsample – number of elements to sample (with replacement) per bootstrap round. If between 0 and 1, treated as a fraction of the number of total number of events (e.g. 0.5 means half the number of events). If not specified, subsampling will not be used and the entire data will be used (without replacement)
- kwargs – all other kwargs will be passed on to the
evaluation_fn
Returns: PermutationImportance.result.ImportanceResultobject which contains the results for each run
Simple Example¶
from sklearn.datasets import load_iris
from sklearn.metrics import accuracy_score
from sklearn.neural_network import MLPClassifier
from PermutationImportance import sklearn_permutation_importance
# Separate out the last 20% for scoring data
iris = load_iris(return_X_y=False)
inputs = iris.get('data')
outputs = iris.get('target')
predictor_names = iris.get('feature_names')
training_inputs = inputs[:int(0.8 * len(inputs))]
training_outputs = outputs[:int(0.8 * len(outputs))]
scoring_inputs = inputs[int(0.8 * len(inputs)):]
scoring_outputs = outputs[int(0.8 * len(outputs)):]
# Train a quick neural net on the data
model = MLPClassifier(solver='lbfgs')
model.fit(training_inputs, training_outputs)
# Package the data into the right shape
scoring_data = (scoring_inputs, scoring_outputs)
# Use the sklearn_permutation_importance to compute importances
result = sklearn_permutation_importance(
model, scoring_data, accuracy_score, 'min', variable_names=predictor_names)
# Get the Breiman-like singlepass results
print("Singlepass")
singlepass = result.retrieve_singlepass()
for predictor in singlepass.keys():
rank, score = singlepass[predictor]
print("Predictor: %s, Rank: %i, Score: %f" % (predictor, rank, score))
# Get the Lakshmanan-like multipass results
print("Multipass")
multipass = result.retrieve_multipass()
for predictor in multipass.keys():
rank, score = multipass[predictor]
print("Predictor: %s, Rank: %i, Score: %f" % (predictor, rank, score))
# Iterate over the (context, result) pairs
for i, (cntxt, res) in enumerate(result):
print("Context %i: %r" % (i, cntxt))
print("Result %i: %r" % (i, res))
Complex Example¶
from sklearn.datasets import load_breast_cancer
from sklearn.ensemble import RandomForestClassifier
from PermutationImportance import sklearn_permutation_importance
from PermutationImportance.metrics import peirce_skill_score
# Separate out the last 20% for scoring data
breast_cancer = load_breast_cancer(return_X_y=False)
inputs = breast_cancer.get('data')
outputs = breast_cancer.get('target')
predictor_names = breast_cancer.get('feature_names')
training_inputs = inputs[:int(0.8 * len(inputs))]
training_outputs = outputs[:int(0.8 * len(outputs))]
scoring_inputs = inputs[int(0.8 * len(inputs)):]
scoring_outputs = outputs[int(0.8 * len(outputs)):]
# Train a quick forest on the data
model = RandomForestClassifier(n_estimators=100, max_depth=4)
model.fit(training_inputs, training_outputs)
# Package the data into the right shape
scoring_data = (scoring_inputs, scoring_outputs)
# ----------- Version to use when only wanting singlepass results --------------
# Use the sklearn_permutation_importance to compute importances
result = sklearn_permutation_importance(
# argmin_of_mean handles bootstrapped metrics
model, scoring_data, peirce_skill_score, 'argmin_of_mean',
variable_names=predictor_names,
# sample (with replacement) 1*(number of samples) 5 times to compute metric distribution
# nbootstrap should typically be 1000, but this is kept small here for printing purposes
nbootstrap=5, subsample=1,
# only perform for the very top predictor (effectively means only compute singlepass results)
nimportant_vars=1)
# Get the Breiman-like singlepass results
print("Singlepass")
singlepass = result.retrieve_singlepass()
for predictor in singlepass.keys():
rank, score = singlepass[predictor]
print("Predictor: %s, Rank: %i, Score: %r" % (predictor, rank, score))
# Get the Lakshmanan-like multipass results
print("Multipass. This should only have 1 item and be not very useful")
multipass = result.retrieve_multipass()
for predictor in multipass.keys():
rank, score = multipass[predictor]
print("Predictor: %s, Rank: %i, Score: %r" % (predictor, rank, score))
# Iterate over the (context, result) pairs
for i, (cntxt, res) in enumerate(result):
print("Context %i: %r" % (i, cntxt))
print("Result %i: %r" % (i, res))
# ------------------------------------------------------------------------------
# ----------- Version to use when wanting multipass results --------------------
# Use the sklearn_permutation_importance to compute importances
result = sklearn_permutation_importance(
# argmin_of_mean handles bootstrapped metrics
model, scoring_data, peirce_skill_score, 'argmin_of_mean',
variable_names=predictor_names,
# sample (with replacement) 1*(number of samples) 5 times to compute metric distribution
# nbootstrap should typically be 1000, but this is kept small here for printing purposes
nbootstrap=5, subsample=1,
nimportant_vars=8) # only perform for the top 8 predictors
# Get the Breiman-like singlepass results
print("Singlepass")
singlepass = result.retrieve_singlepass()
for predictor in singlepass.keys():
rank, score = singlepass[predictor]
print("Predictor: %s, Rank: %i, Score: %r" % (predictor, rank, score))
# Get the Lakshmanan-like multipass results
print("Multipass. This should have exactly 8 items")
multipass = result.retrieve_multipass()
for predictor in multipass.keys():
rank, score = multipass[predictor]
print("Predictor: %s, Rank: %i, Score: %r" % (predictor, rank, score))
# Iterate over the (context, result) pairs
for i, (cntxt, res) in enumerate(result):
print("Context %i: %r" % (i, cntxt))
print("Result %i: %r" % (i, res))
# ------------------------------------------------------------------------------
# Use the plotting code in examples/plotting.py, found here:
# https://github.com/gelijergensen/PermutationImportance
try:
from plotting import plot_variable_importance
except Exception as e:
print("An error occurred while plotting. You probably don't have matplotlib installed")
print(e)
else:
plot_variable_importance(
result, 'example_singlepass_permutation.png', multipass=False)
plot_variable_importance(
result, 'example_multipass_permutation.png', multipass=True)
Method-Specific¶
-
PermutationImportance.permutation_importance.permutation_importance(scoring_data, scoring_fn, scoring_strategy, variable_names=None, nimportant_vars=None, njobs=1)[source] Performs permutation importance over data given a particular set of functions for scoring and determining optimal variables
Parameters: - scoring_data – a 2-tuple
(inputs, outputs)for scoring in thescoring_fn - scoring_fn – a function to be used for scoring. Should be of the form
(training_data, scoring_data) -> some_value - scoring_strategy – a function to be used for determining optimal
variables. Should be of the form
([some_value]) -> index - variable_names – an optional list for variable names. If not given, will use names of columns of data (if pandas dataframe) or column indices
- nimportant_vars – number of variables to compute multipass importance for. Defaults to all variables
- njobs – an integer for the number of threads to use. If negative, will
use
num_cpus + njobs. Defaults to 1
Returns: PermutationImportance.result.ImportanceResultobject which contains the results for each run- scoring_data – a 2-tuple
Example¶
from sklearn.datasets import load_iris
from sklearn.metrics import accuracy_score
from sklearn.neural_network import MLPClassifier
from PermutationImportance import permutation_importance
# Separate out the last 20% for scoring data
iris = load_iris(return_X_y=False)
inputs = iris.get('data')
outputs = iris.get('target')
predictor_names = iris.get('feature_names')
training_inputs = inputs[:int(0.8 * len(inputs))]
training_outputs = outputs[:int(0.8 * len(outputs))]
scoring_inputs = inputs[int(0.8 * len(inputs)):]
scoring_outputs = outputs[int(0.8 * len(outputs)):]
# Some model we are interested in
model = MLPClassifier(solver='lbfgs')
model.fit(training_inputs, training_outputs)
def score_model(training_data, scoring_data):
"""Custom function to use for scoring. Notice that we are using a global
model here, rather than just reassemble the model each time
:param training_data: should be ignored for permutation importance
:param scoring_data: (scoring_inputs, scoring_outputs)
"""
scoring_ins, scoring_outs = scoring_data
return accuracy_score(scoring_outs, model.predict(scoring_ins))
# Package the data into the right shape
scoring_data = (scoring_inputs, scoring_outputs)
# Use the permutation_importance to compute importances
result = permutation_importance(
scoring_data, score_model, 'min', variable_names=predictor_names)
# Get the Breiman-like singlepass results
print("Singlepass")
singlepass = result.retrieve_singlepass()
for predictor in singlepass.keys():
rank, score = singlepass[predictor]
print("Predictor: %s, Rank: %i, Score: %f" % (predictor, rank, score))
# Get the Lakshmanan-like multipass results
print("Multipass")
multipass = result.retrieve_multipass()
for predictor in multipass.keys():
rank, score = multipass[predictor]
print("Predictor: %s, Rank: %i, Score: %f" % (predictor, rank, score))
# Iterate over the (context, result) pairs
for i, (cntxt, res) in enumerate(result):
print("Context %i: %r" % (i, cntxt))
print("Result %i: %r" % (i, res))